
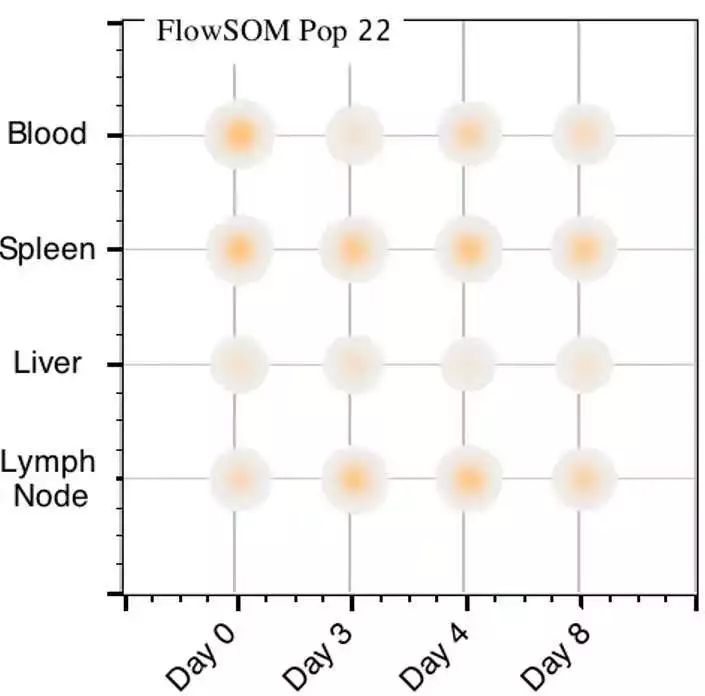
Serena shows how each of these 10 sub-populations can then be interrogated to reveal unique phenotypes enabling further focussed characterisation experiments. Instead, supplementary markers within a larger panel reveal 10 unique and discrete sub-clusters of Tregs. Using Tregs as a representative study population, Serena demonstrates how FlowJo™ goes beyond conventional approaches that limit analysis to classic core phenotyping markers of the population: CD3+ CD4+ CD25+ CD127. Serena then presents a simple 4 step FlowJo™ workflow to perform a quality assessment and control (QC) of your data, collectively visualize all the cells simultaneously identify the presence of any novel sub-populations present within your study sample and finally conduct a detailed characterization of those specific subsets of interest to you. Frank starts by highlighting common challenges and limitations of conventional 2D sequential gating – the common approach to analysing flow data. Multiparameter flow cytometry data analysis done the customary way: bi-dimensional XY plots that gate on a gate on a gate etc followed by a practical demonstration of a more powerful and much easier way to simultaneously analyse multiple parameters using FlowJo™ plugins.įrank and Serena demonstrate how you too could gain deeper biological insights by leveraging an easy-to-apply approach to higher dimension data analysis.


 0 kommentar(er)
0 kommentar(er)
